Help Page
Documentation and usage instructions
Visit our YouTube video to learn how to use it and how to evaluate the results.
-Fundamentals of ARTS and FunARTS Pipeline-
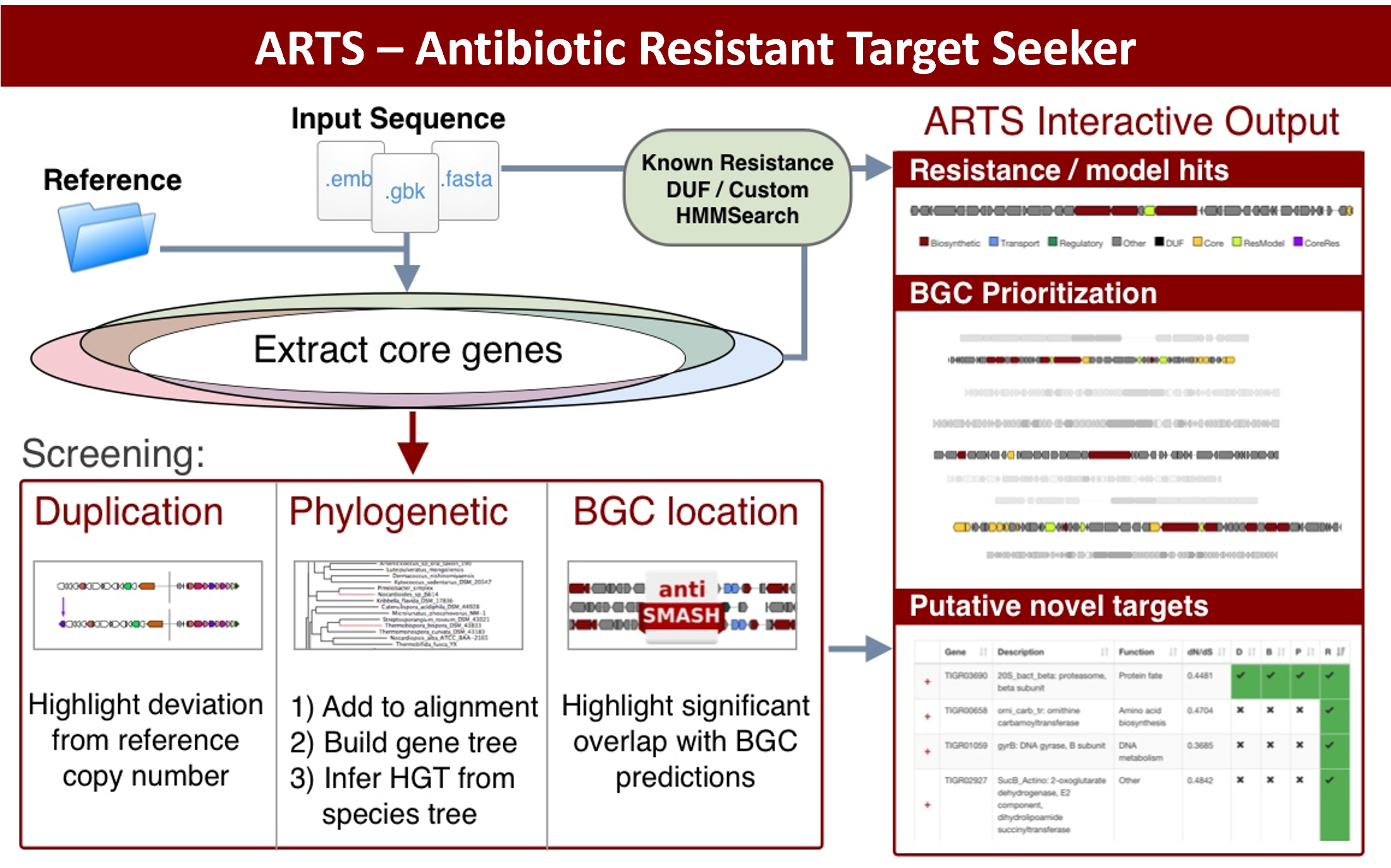
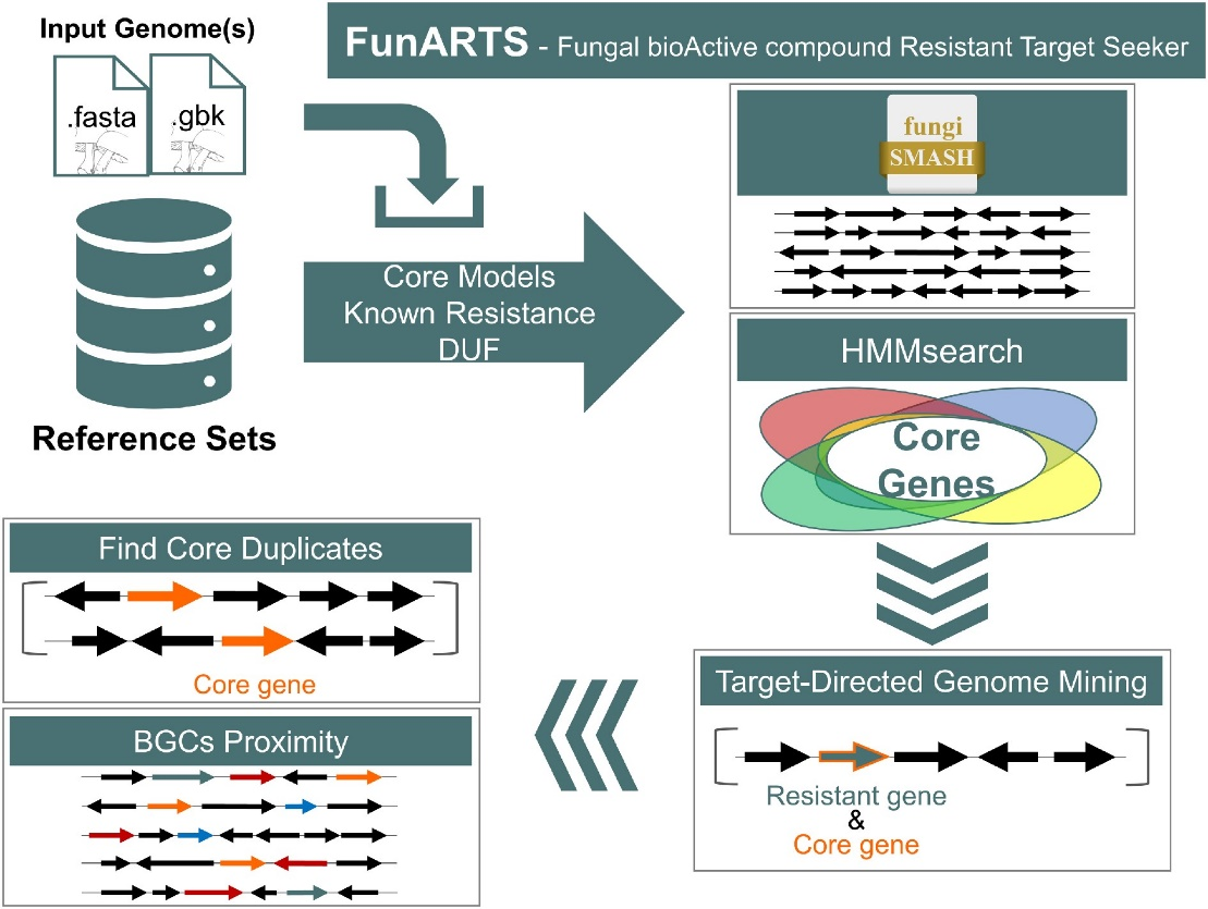
Understanding ARTS and FunARTS results
The goals of this tool are to automate the process of performing target directed genome mining (TDGM), search for potential novel antibiotic targets and prioritize putative secondary metabolite gene clusters.
There are three fundamental criteria for TDGM as listed below:
- Proximity check: Cross reference locations with secondary metabolite gene clusters. For more information about BGC prediction, please visit the antiSMASH help page.
- Uncommon duplication check: Highlight potential repurposed primary metabolism genes.
- Phylogenetic incongruence: Highlight essential genes with evidence of inter-genus horizontal transfer.
Please visit the ARTS or FunARTS help pages for more information, or our YouTube channel for tutorials.
-Submitting a Query-
Selection of genome pool and ARTS/FunARTS criterion:
- First select the genome pool for searching. This selects the target taxonomic domain (Bacteria or Fungus) to define the scope of the dataset.
- Select one of the fundamental TDGM criteria for specialized queries. Note that you can search for HGT evidence via selecting the respective search option from "Core Hits" or "Summary Results" under Bacteria.
-
After selecting main and sub categories, add available search option terms of your
choice and provide a string (e.g.
streptomyces) or a substring (e.g.strepto) to query the database.
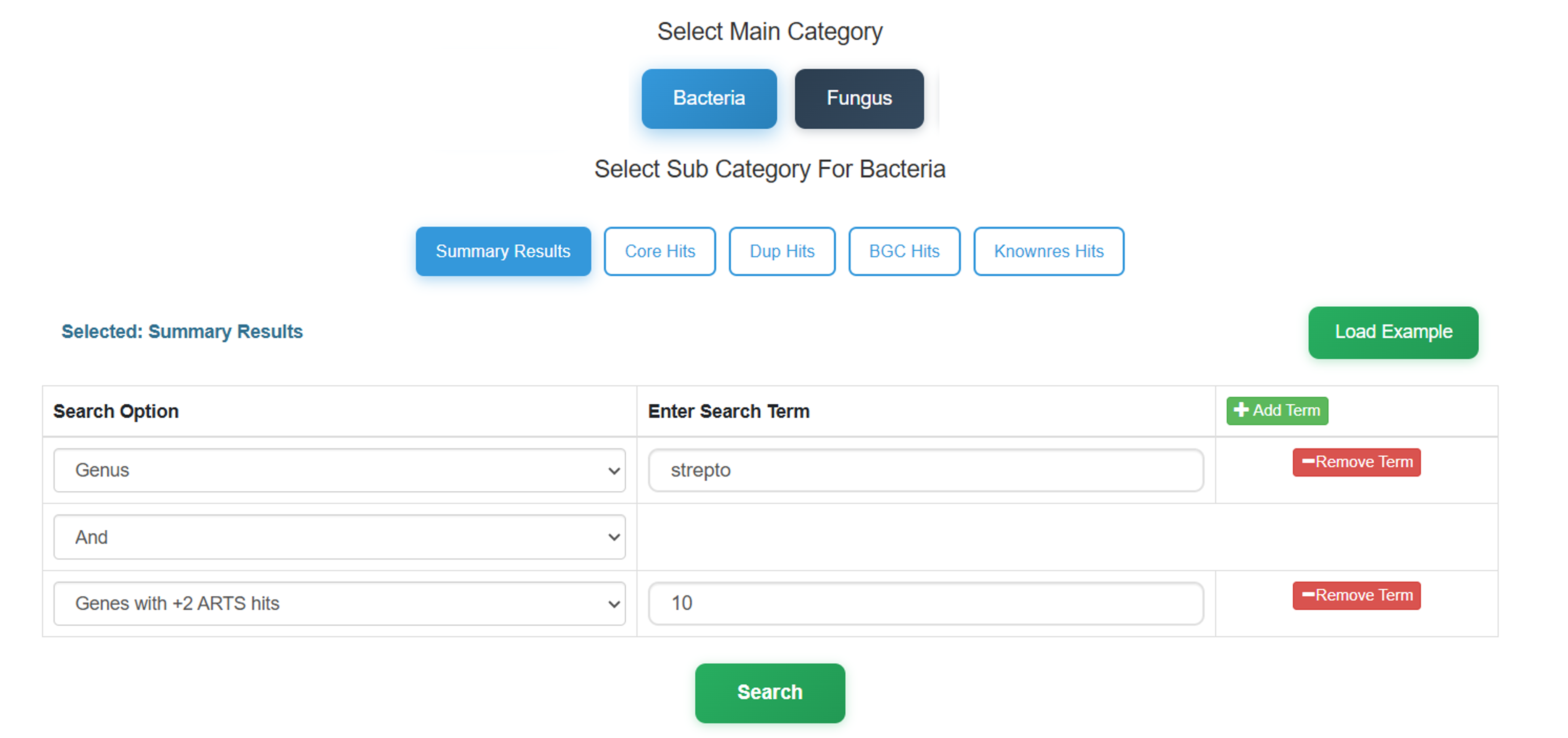
Exploring Results
Your applied search query will lead you to a result page, consisting of a dynamic table where you can search for your specific keywords throughout the table, download your selection, or navigate to detailed results or other databases by clicking on provided link-outs shown in blue text.
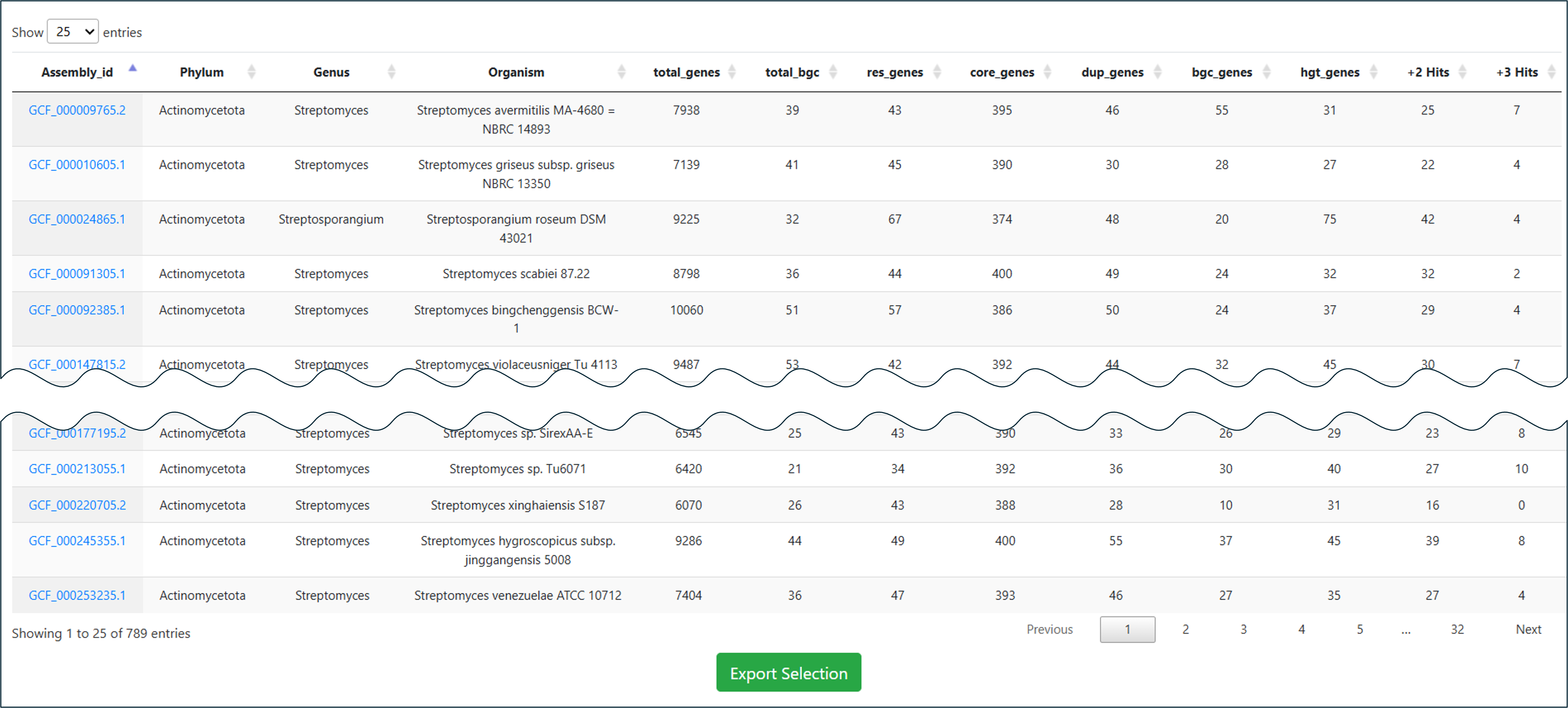
Case Study 1 – Searching Core Hits for Specific Resistance Candidate
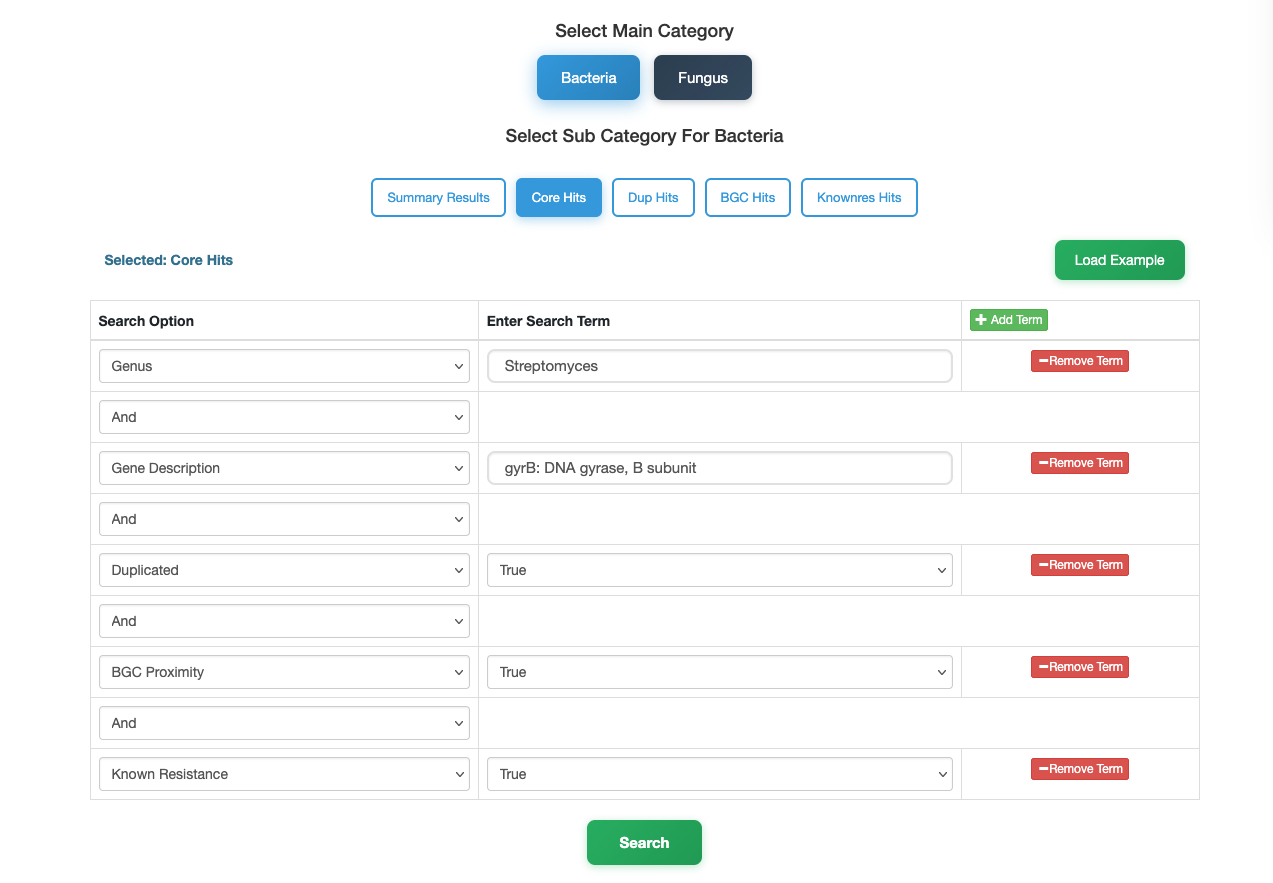
In this case, we demonstrate how to find high-priority drug targets within the genus Streptomyces using the DNA gyrase subunit B (gyrB) as an example. Follow these steps to filter thousands of genomes down to the most promising candidates:
- Select Category: Navigate to the Bacteria database and choose the Core Hits sub-category to screen for essential genes.
- Define Organism: In the search panel, set the Genus to
Streptomyces. - Specify Target: Use the Add Term button to add a new filter. Set Description to
gyrB(or "DNA gyrase subunit B"). -
Apply Resistance Filters: To identify targets with strong resistance potential, add the following terms and set them all to
True:- Duplicated – Is the gene duplicated?
- Proximity – Is it near a biosynthetic gene cluster?
- Known Resistance – Does it match known resistance factors?
- Search: Click the Search button.
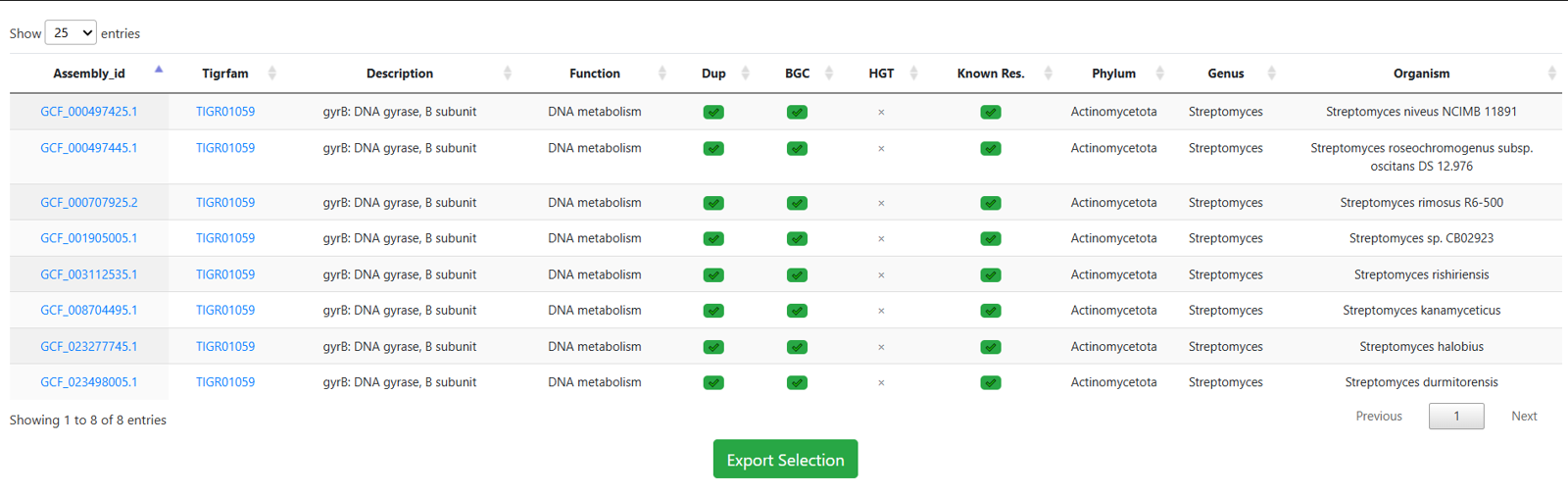
Result: This stringent multi-parametric query reduces the broad dataset to a small number of specific strains, allowing you to focus on the most relevant targets immediately.
Case Study 2 – Identifying Resistance-Linked BGCs (BGC Hits)
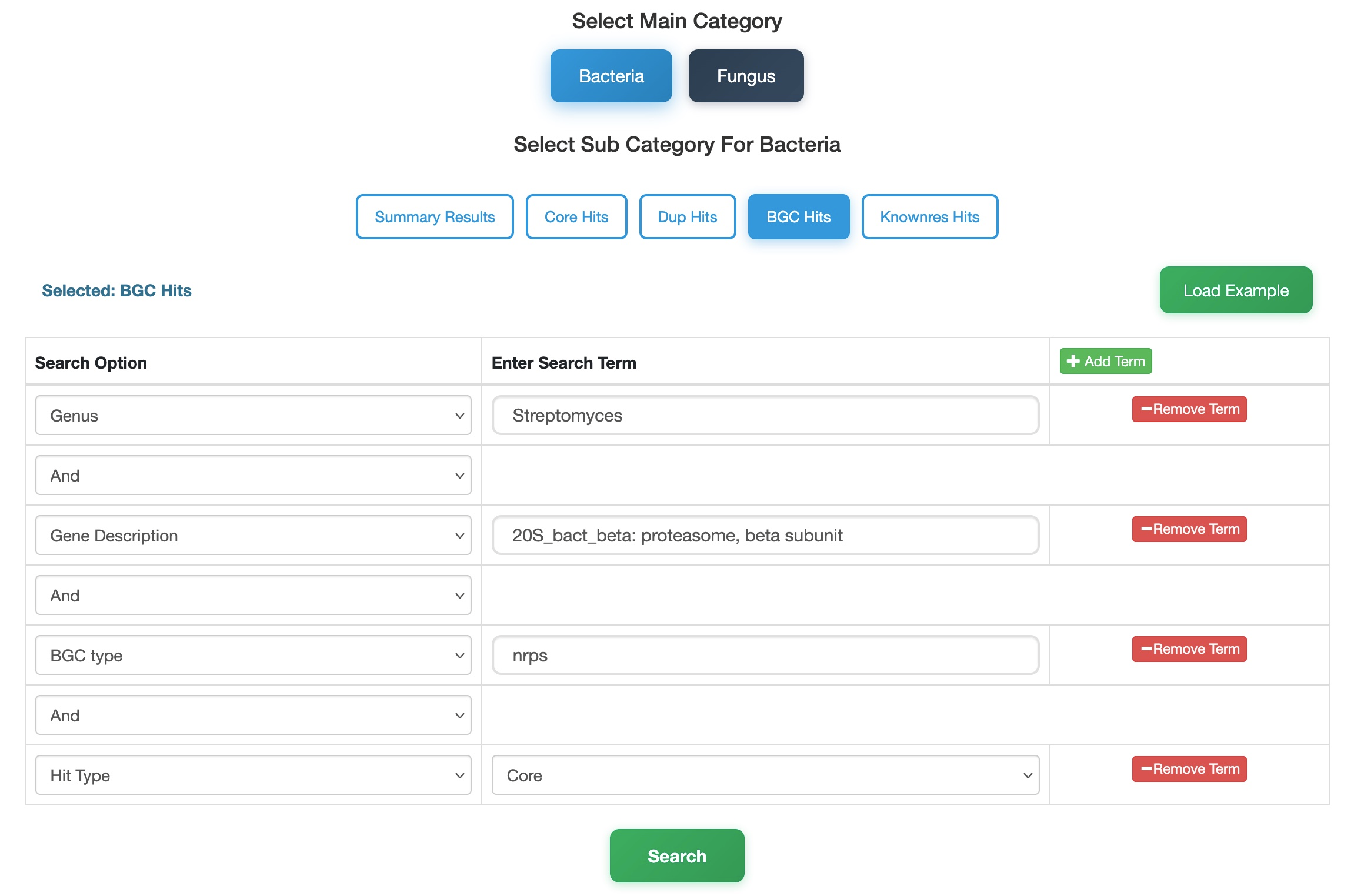
In this tutorial, we use the BGC Hits category to locate β-proteasome subunit genes specifically annotated as essential core genes within Non-Ribosomal Peptide Synthetase (NRPS) clusters in Streptomyces.
- Select Database & Category: Navigate to the Bacteria database and choose the BGC Hits sub-category. This category is ideal for linking specific targets to defined biosynthetic cluster types.
- Define Organism: In the search panel, set the Genus to
Streptomyces. - Specify Target: Use the Add Term button to add a description filter. Set Description to
20S_bact_beta: proteasome, beta subunit. - Filter by Biosynthetic Type: To narrow down the search to a specific cluster class, set the BGC Type to
NRPS. - Define Hit Nature: Set the Hits Type to
Core. SelectingCoreensures the gene is predicted as an essential core gene. Alternatively, selectingResModelwould identify co-localized resistance genes. - Search: Click the Search button.
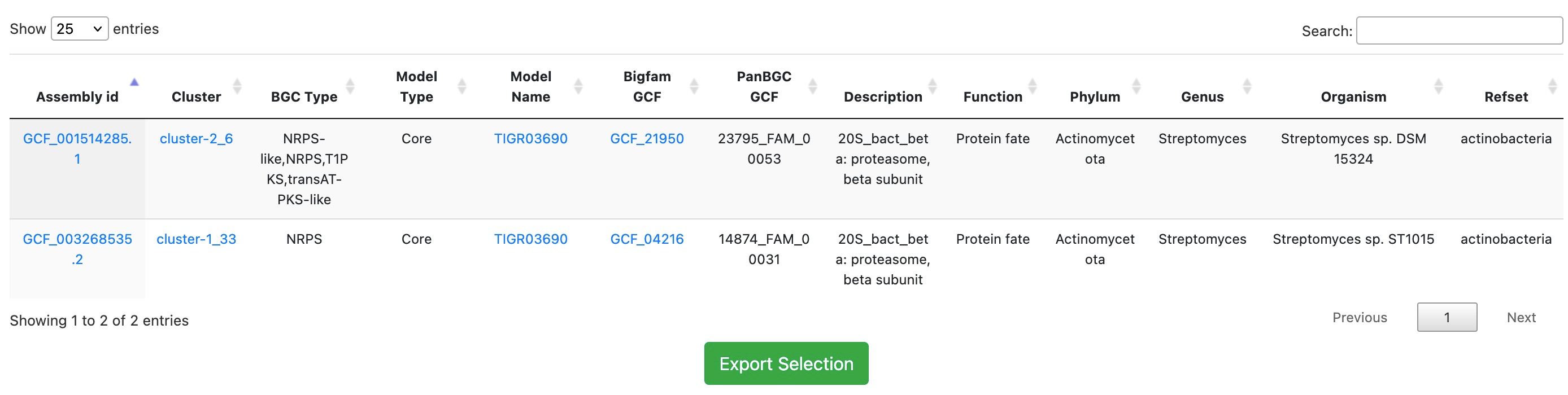
Result: The search returns only those BGCs containing the beta-proteasome subunit gene as an essential component within NRPS clusters, effectively filtering out unrelated cluster types.
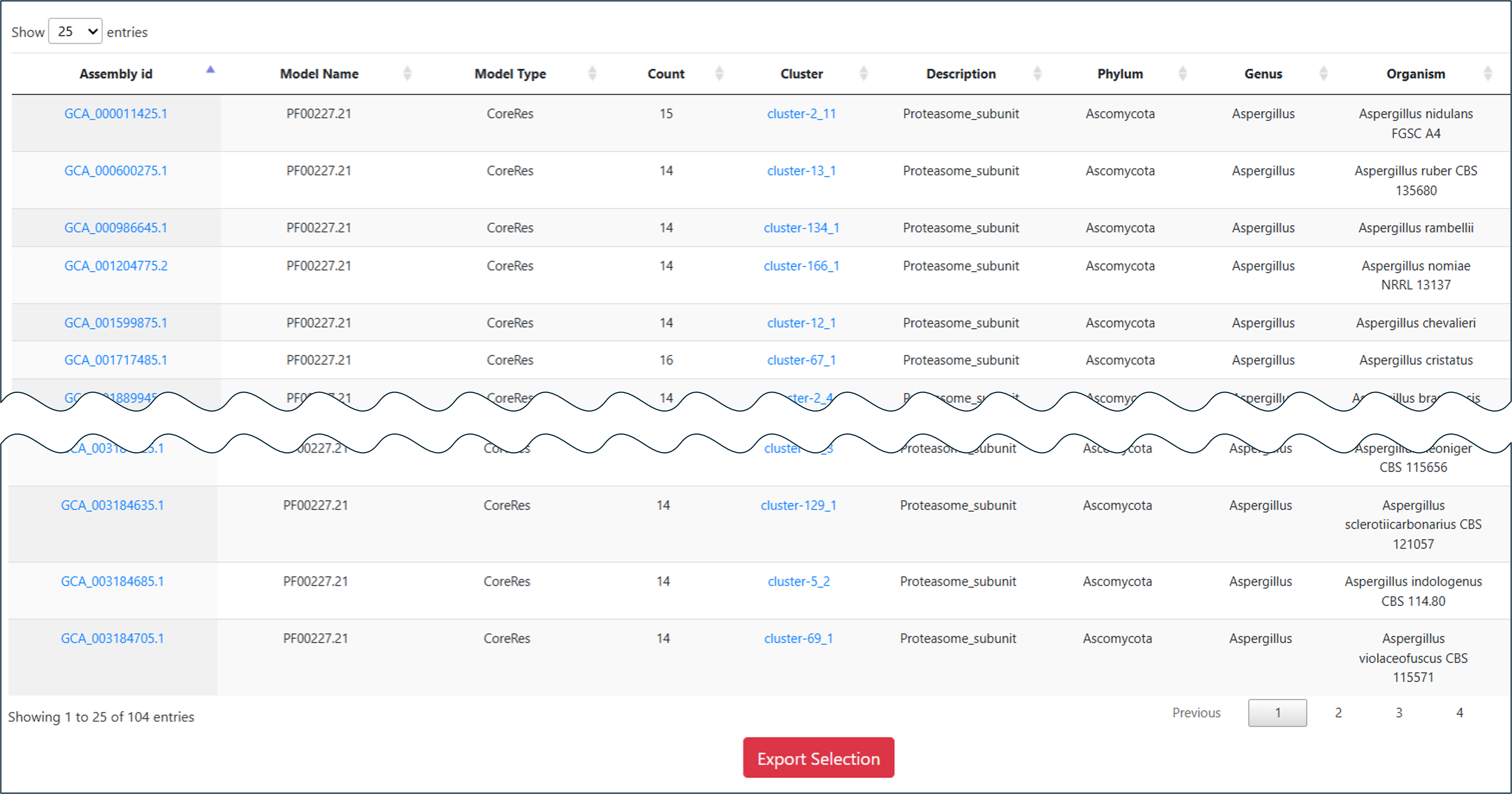
Links from the Assembly id column take you to the corresponding genome result. The Cluster column leads to the antiSMASH result page, Model Name leads to phylogenetic and statistical information about the selected model throughout ARTS-DB 2.0, and BiGFAM GCF and PanBGC GCF link to the associated Gene Cluster Families via BiGFAM-DB and PanBGC-DB, respectively.
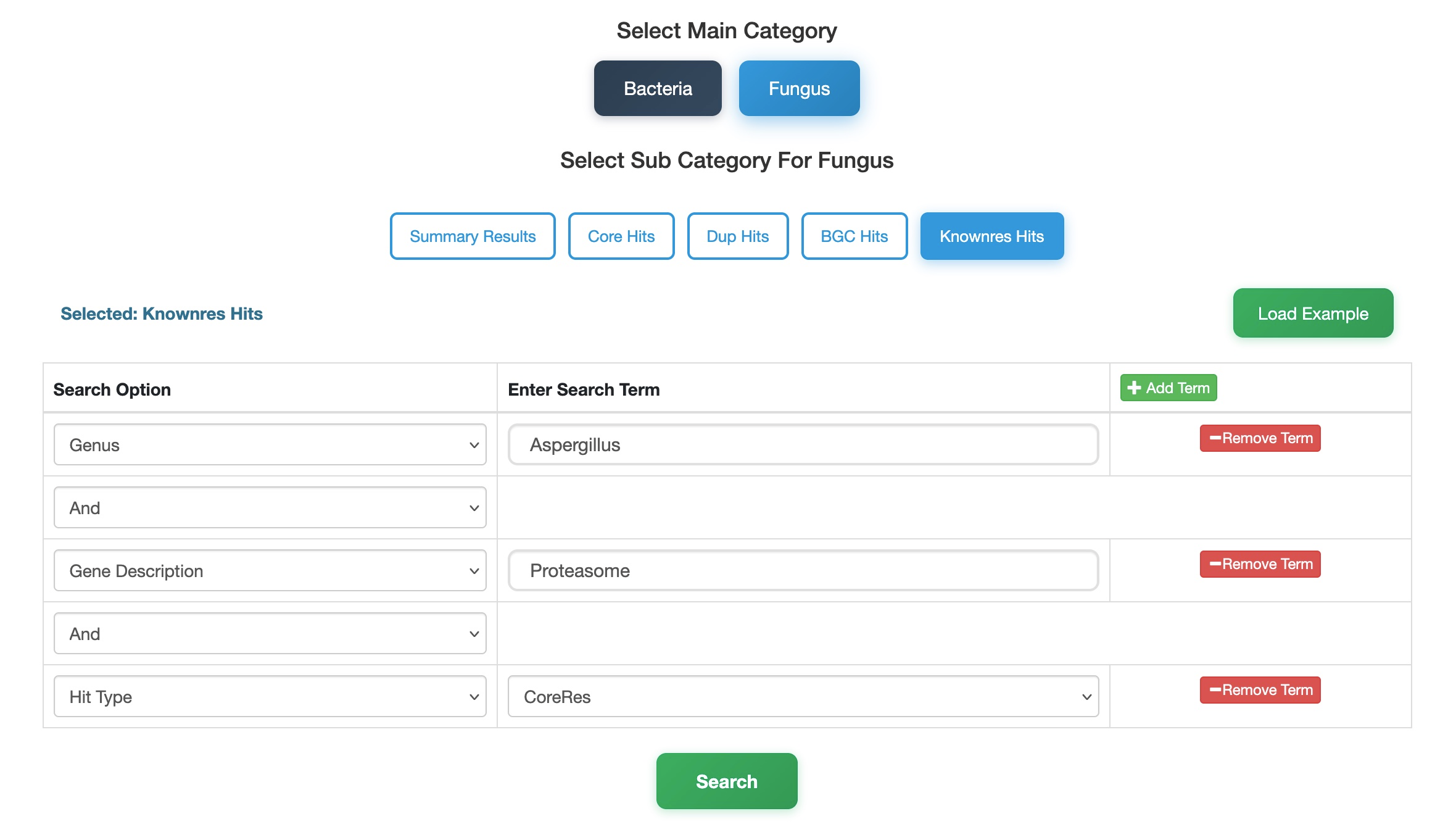
Case Study 3 – Fungal Resistance Exploration
Here, we explore fungal resistance mechanisms by searching for proteasome subunits co-localized with essential genes within the genus Aspergillus.
- Select Database & Category: Navigate to the Fungi database and select the KnownRes Hits sub-category. This module is specifically designed to query resistance determinants based on their genomic context.
- Define Organism: In the search panel, set the Genus to
Aspergillus. - Specify Target: Use the Add Term button to add a description filter. Set Description to
proteasome. - Refine Resistance Type: To focus on resistance genes that map to the same locus as an essential core gene (co-localization), add the Hit Type filter and set it to
CoreRes. You can also chooseResModelfor BGC-located genes orResGenefor standalone resistance genes. - Search: Click the Search button.
Result: This targeted structured query effectively filters the fungal dataset, retrieving Aspergillus entries that satisfy strict co-localization criteria and identify precise resistance mechanisms.
-Model-Oriented Search-
Searching Potential Targets
Here you can provide a substring of gene description and select a gene (classified from
TIGRFAM and BUSCO models) from suggestions. If you have any specific models in mind
(e.g.:TIGR01059), you can also pass that as a query and if the model exists in
ARTS-DB, you will be directed to the result page.
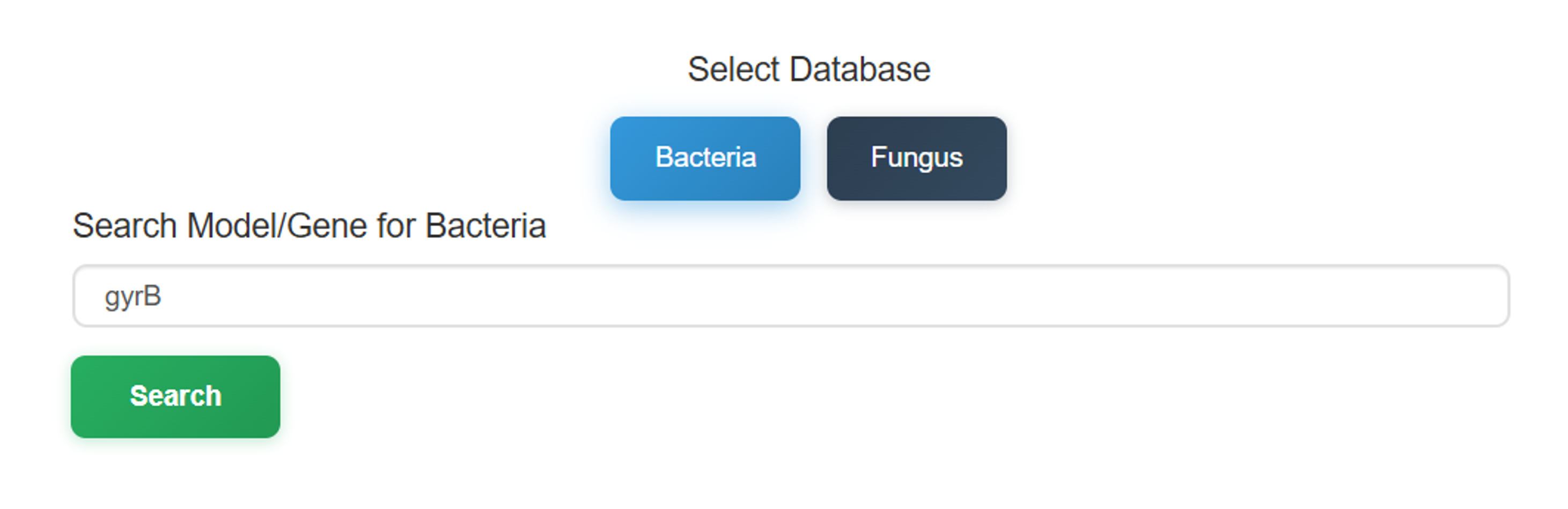
The Model-Oriented Search begins with a taxonomic selection between Bacteria and Fungus.
The process then continues as follows: enter or select the desired gene name in the search
bar (for example, gyrB) and click the Search button.
The Model-Oriented result page (Example) consists of phylogenetical and statistical information about the selected gene in terms of ARTS/FunARTS criteria hits such as:
- Distribution of BGC Proximity Hits: Here you can investigate target genes affinity to be proximate to certain types of biosynthetic gene clusters or the taxonomic distribution of resulting BGC proximity hits for that gene.
- Distribution of Duplication Hits: Here the duplication rates of the selected gene with respect to a specific phylum can be investigated. This is also important for BGC expression studies since repurposing primary metabolites can help boost secondary metabolite production.
- Phylogenetic Distribution of target gene with Multiple ARTS Hits: Here you can investigate your targets affinity to be a likely target for a resistance mechanism in corresponding phyla. Please note that these plots are generated for presenting a broader sense of the "resistance potential" of gene of interest. Further investigation should be done by executing specific queries in order to narrow down potential targets in accordance with your current research.
- External Resources: Here you can further check the link outs to investigate your gene of interests affiliation with databases such as UniProt, MIBiG, ChEMBL, DrugBank and DrugCentral.